14.3 Transcription: From Gene to Message
Elizabeth Dahlhoff
Learning Objectives
By the end of this section, you will be able to do the following:
- Explain the role of promoters and RNA polymerase in transcription of DNA to messenger RNA.
- Explain the significance of transcription factors in eukaryotic transcription.
- Describe the major steps of transcription, including the roles of template strand, reading strand, and the directionality of the growing mRNA.
- Describe how transcription is terminated.
Transcription
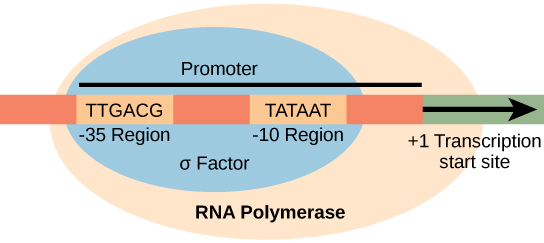
Transcription differs between prokaryotes (single-celled organisms that lack membrane-bound nuclei and membrane-enclosed organelles) and eukaryotes (organized around histone proteins), but the fundamentals are the same. Transcription of a gene begins when the enzyme RNA polymerase binds to DNA at the promoter region of that gene. A promoter is a DNA sequence onto which the transcription machinery, including RNA polymerase, binds and initiates transcription. Promoters exist a certain distance upstream of the genes they regulate.
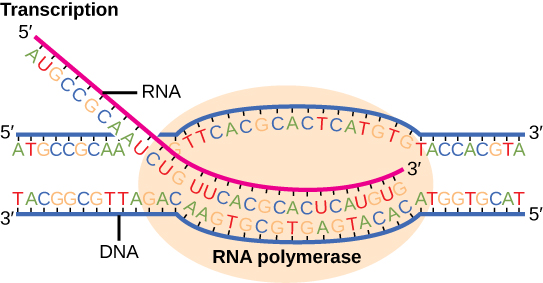
Transcription requires the DNA double helix to partially unwind in the region of messenger RNA (mRNA) synthesis. The region of unwinding is called a transcription bubble. Transcription always proceeds from the same DNA strand for each gene, which is called the template strand. The mRNA product is complementary and antiparallel to the template strand and is almost identical to the other DNA strand, called the coding strand. There are two differences: 1) in mRNA, all of the T nucleotides are replaced with U nucleotides; 2) in mRNA, the sugar phosphate “backbone” of the molecule is ribose, whereas for DNA, the sugar is deoxyribose. In an mRNA-DNA strand pairing during transcription, A can bind U via two hydrogen bonds, just as in A–T pairing in a DNA double helix. Messenger RNA is always synthesized in the 5′ to 3′ direction.
Once a gene is transcribed, the prokaryotic polymerase needs to be instructed to dissociate from the DNA template and liberate the newly made mRNA. Depending on the gene being transcribed, there are two kinds of termination signals. One is protein-based and the other is RNA-based. Rho-dependent termination is controlled by the rho protein, which tracks along behind the polymerase on the growing mRNA chain. Near the end of the gene, the polymerase encounters a run of G nucleotides on the DNA template and it stalls. As a result, the rho protein collides with the polymerase. The interaction with rho releases the mRNA from the transcription bubble.
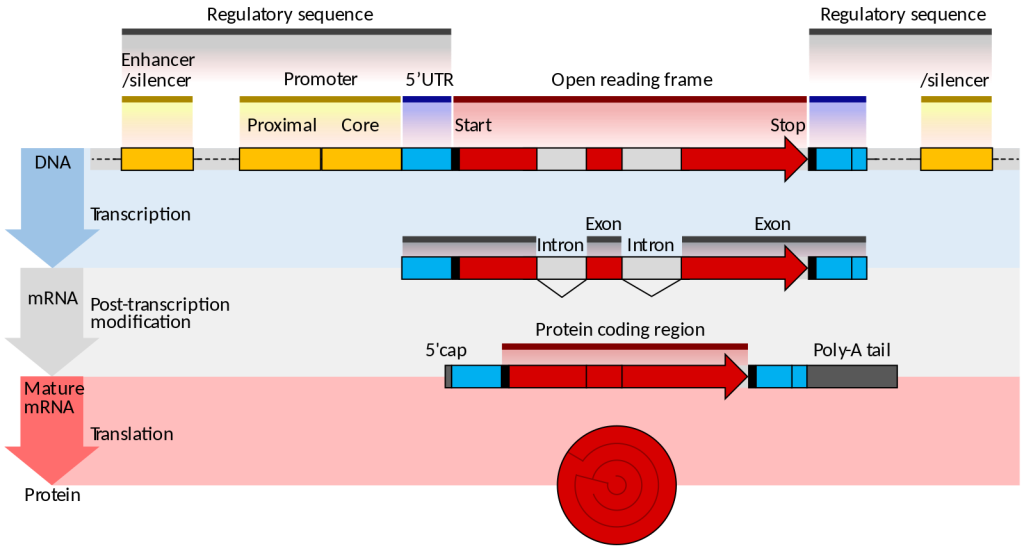
Unlike prokaryotes, eukaryotes have multiple linear chromosomes. Eukaryotic mRNAs typically specify a single protein, as opposed to the mRNA transcribed from multi-gene units called operons. The structure of a generalized eukaryotic protein-coding gene and its product is shown below.
Prokaryotes and eukaryotes perform fundamentally the same process of transcription, with a few key differences. The most important difference between prokaryote and eukaryote transcription is due to the latter’s membrane-bound nucleus and organelles. With the genes bound in a nucleus, the eukaryotic cell must be able to transport its mRNA to the cytoplasm and must protect its mRNA from degrading before it is translated. Eukaryotes also employ three different polymerases that each transcribe a different subset of genes.
| RNA Polymerase | Cellular Compartment | Product of Transcription |
|---|---|---|
| I | Nucleolus | All rRNAs except 5S rRNA |
| II | Nucleus | All protein-coding nuclear pre-mRNAs and most small nuclear RNAs |
| III | Nucleus | 5S rRNA, tRNAs, and some small nuclear RNAs |
RNA polymerase II is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription but before translation. For clarity, we will only use the term “mRNAs” to describe only the mature, processed molecules that are ready to be translated. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes.
RNA Polymerase II Promoters and Transcription Factors
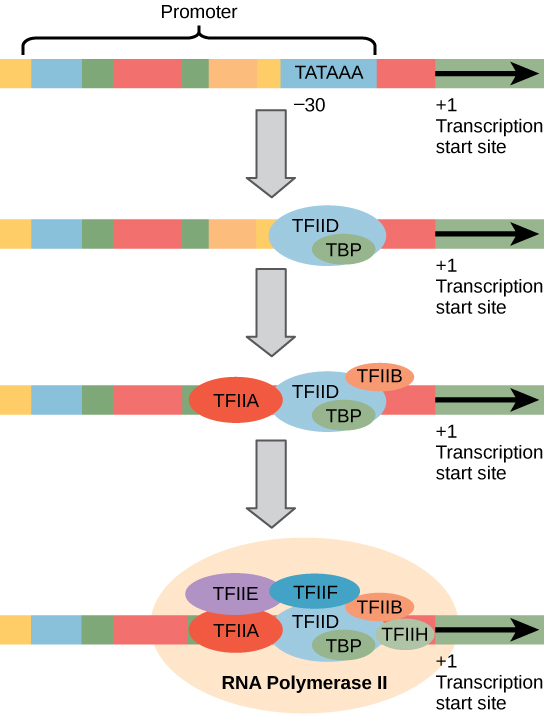
Eukaryotic promoters are much larger and more intricate than prokaryotic promoters. However, both have a sequence similar to the -10 sequence of prokaryotes. In eukaryotes, this sequence is called the TATA box, and has the consensus sequence TATAAA on the coding strand. It is located at -25 to -35 bases relative to the initiation (+1) site.
Like prokaryotic cells, the transcription of genes in eukaryotes requires the action of an RNA polymerase to bind to a DNA sequence upstream of a gene in order to initiate transcription of that gene. However, unlike prokaryotic cells, the eukaryotic RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. RNA polymerase by itself cannot initiate transcription in eukaryotic cells.
Here is a nice visualization (Video 14.3.1) of the initiation of RNA transcription and how the long messenger RNA chain is synthesized.
Video 14.3.1. DNA Transcription (Advanced) by DNA Learning Center
Eukaryotic Elongation and Termination
Following the formation of the pre-initiation complex, the polymerase is released from the promoter, and elongation is allowed to proceed as it does in prokaryotes, with the polymerase synthesizing pre-mRNA in the 5′ to 3′ direction. As discussed previously, RNA polymerase II transcribes the major share of eukaryotic genes, so in this section we will focus on how this polymerase accomplishes elongation and termination.
Although the enzymatic process of elongation is essentially the same in eukaryotes and prokaryotes, the DNA template is considerably more complex. When eukaryotic cells are not dividing, their genes exist as a diffuse mass of DNA and proteins called chromatin. The DNA is tightly packaged around charged histone proteins at repeated intervals. These DNA–histone complexes, collectively called nucleosomes, are regularly spaced and include 146 nucleotides of DNA wound around eight histones like thread around a spool. For polynucleotide synthesis to occur, the transcription machinery needs to move histones out of the way every time it encounters a nucleosome. This is accomplished by a special protein complex called FACT, which stands for “facilitates chromatin transcription.” This complex pulls histones away from the DNA template as the polymerase moves along it. Once the pre-mRNA is synthesized, the FACT complex replaces the histones to recreate the nucleosomes.
The termination of transcription is different for the different polymerases. Unlike in prokaryotes, elongation by RNA polymerase II in eukaryotes, which is the RNA polymerase responsible for protein synthesis, takes place 1,000 to 2,000 nucleotides beyond the end of the gene being transcribed. This pre-mRNA tail is subsequently removed by cleavage during mRNA processing.
Regulation of Transcription
The DNA of prokaryotes is organized into a circular chromosome. Proteins that are needed for a specific function, or that are involved in the same biochemical pathway, are often encoded together in blocks called operons. For example, all of the genes needed to use lactose as an energy source are coded next to each other in the lactose (or lac) operon, and transcribed into a single mRNA. You will learn more about operons and other aspects of prokaryotic and eukaryotic gene expression in BIOL 1C.
Practice Questions
Section Summary
In prokaryotes, mRNA synthesis is initiated at a promoter sequence on the DNA template comprising two consensus sequences that recruit RNA polymerase. Elongation synthesizes mRNA in the 5′ to 3′ direction. Termination liberates the mRNA and occurs either by rho protein interaction or by the formation of an mRNA hairpin. Transcription in eukaryotes involves one of three types of polymerases, depending on the gene being transcribed. RNA polymerase II transcribes all of the protein-coding genes, whereas RNA polymerase I transcribes rRNA genes, and RNA polymerase III transcribes rRNA, tRNA, and small nuclear RNA genes. The initiation of transcription in eukaryotes involves the binding of several transcription factors to complex promoter sequences that are usually located upstream of the gene being copied. The mRNA is synthesized in the 5′ to 3′ direction, and the FACT complex moves and reassembles nucleosomes as the polymerase passes by. Whereas RNA polymerases I and III terminate transcription by protein- or RNA hairpin-dependent methods, RNA polymerase II transcribes for 1,000 or more nucleotides beyond the gene template and cleaves the excess during pre-mRNA processing, which is discussed in the following chapter.
Glossary
- consensus
- DNA sequence that is used by many species to perform the same or similar functions
- core enzyme
- prokaryotic RNA polymerase consisting of α, α, β, and β‘ but missing σ; this complex performs elongation
- downstream
- nucleotides following the initiation site in the direction of mRNA transcription; in general, sequences that are toward the 3′ end relative to a site on the mRNA
- hairpin
- structure of RNA when it folds back on itself and forms intramolecular hydrogen bonds between complementary nucleotides
- initiation site
- nucleotide from which mRNA synthesis proceeds in the 5′ to 3′ direction; denoted with a “+1”
- nontemplate (coding) strand
- strand of DNA that is not used to transcribe mRNA; this strand is identical to the mRNA except that T nucleotides in the DNA are replaced by U nucleotides in the mRNA
- promoter
- DNA sequence to which RNA polymerase and associated factors bind and initiate transcription
- TATA box
- conserved promoter sequence in eukaryotes and prokaryotes that helps to establish the initiation site for transcription
- template strand
- strand of DNA that specifies the complementary mRNA molecule
- transcription bubble
- region of locally unwound DNA that allows for transcription of mRNA
- upstream
- nucleotides preceding the initiation site; in general, sequences toward the 5′ end relative to a site on the mRNA
Video 14.3.2. The Central Dogma of Biology by DNA Learning Center
Figure Descriptions
Figure 14.3.1. The diagram illustrates how the σ subunit of prokaryotic RNA polymerase recognizes specific consensus sequences in the promoter region upstream of the transcription start site. The DNA strand contains two highlighted regions: the -35 region labeled “TTGACG” and the -10 region labeled “TATAAT.” These regions lie within a blue oval labeled “σ Factor” and are part of a larger orange oval labeled “RNA Polymerase.” A black arrow points to the right from the +1 transcription start site, representing the direction of transcription. The -35 and -10 regions are shown within the promoter sequence, which the σ subunit binds to before transcription begins. [Return to Figure 14.3.1]
Figure 14.3.2. The illustration shows a close-up of transcription. Two strands of DNA are partially unwound inside an orange oval representing RNA polymerase. The bottom strand is the DNA template, and the top strand is the coding strand. A pink RNA strand is forming by base pairing with the template strand, substituting uracil (U) for thymine (T). The RNA is synthesized in the 5′ to 3′ direction, with the 5′ end emerging from the polymerase. DNA ahead of the polymerase is unwound, while DNA behind it rewinds into a double helix. Labels identify RNA, DNA, and RNA polymerase, with nucleotide sequences in color-coded letters for adenine (A), thymine (T), cytosine (C), guanine (G), and uracil (U). [Return to Figure 14.3.2]
Figure 14.3.3. The diagram illustrates the structure and expression of a eukaryotic protein-coding gene in three stages. At the DNA level, yellow enhancer and silencer regions flank a promoter containing proximal and core segments. The promoter precedes the start site of the red protein-coding region, which includes alternating white introns and red exons, ending at a stop site before another enhancer/silencer. During transcription, the pre-mRNA mirrors this structure. In post-transcriptional modification, introns are removed, and a blue 5′ cap is added at the start while a poly-A tail is added at the end. The mature mRNA has blue 5′ and 3′ untranslated regions, a continuous red protein-coding sequence, and is translated into a protein. [Return to Figure 14.3.3]
Figure 14.3.4. The diagram depicts four steps in forming the transcription initiation complex at a eukaryotic promoter transcribed by RNA polymerase II. At the top, a DNA segment includes a labeled promoter region with a TATA box positioned about 30 bases upstream of the +1 transcription start site. In the first step, transcription factor TFIID, containing the TBP (TATA-binding protein), binds the TATA box. Next, TFIIA and TFIIB join the complex. In the final step, additional transcription factors TFIIE, TFIIF, and TFIIH assemble with RNA polymerase II at the promoter, forming the complete transcription initiation complex. Arrows indicate the direction of transcription from the +1 site. [Return to Figure 14.3.4]
Licenses and Attributions
Media Attributions
- Prokaryote promoter © OpenStax is licensed under a CC BY (Attribution) license
- mRNA © OpenStax is licensed under a CC BY (Attribution) license
- Gene_structure_eukaryote_2_annotated.svg © Thomas Shafee is licensed under a CC BY (Attribution) license
- Eukaryote Transcription factors © OpenStax is licensed under a CC BY (Attribution) license

Although there is a large variety, each item of food in the image above ultimately can be linked back to photosynthesis in its own way. Meats and dairy link, because the animals were fed plant-based foods, such as the soybeans that both make up tofu and feed the cows that become beef. The breads, lentils, and rice come largely from starchy grains, which are the seeds of photosynthesis-dependent plants. Even dessert and drinks contain sugars like sucrose, a plant product, a disaccharide, a carbohydrate molecule, which is built directly from photosynthesis. Moreover, less obvious products such as paper goods are generally plant products, and many plastics (abundant as products and packaging) are derived from “algae” (unicellular plant-like organisms, and cyanobacteria). Virtually every spice and flavoring in our meals was produced by a plant as a leaf, root, bark, flower, fruit, or stem. Ultimately, photosynthesis connects to every meal and every food a person consumes.
