20.4 Response to the Signal
Learning Objectives
By the end of this section, you will be able to do the following:
- Describe how signaling pathways direct protein expression, cellular metabolism, and cell growth.
- Describe the role of apoptosis in the development and maintenance of a healthy organism.
Cells respond to the activation of receptors by their ligands in a variety of different ways. The results of signaling pathways are extremely varied and depend on the type of cell involved as well as the external and internal conditions. A small sampling of responses is described below.
Gene Expression
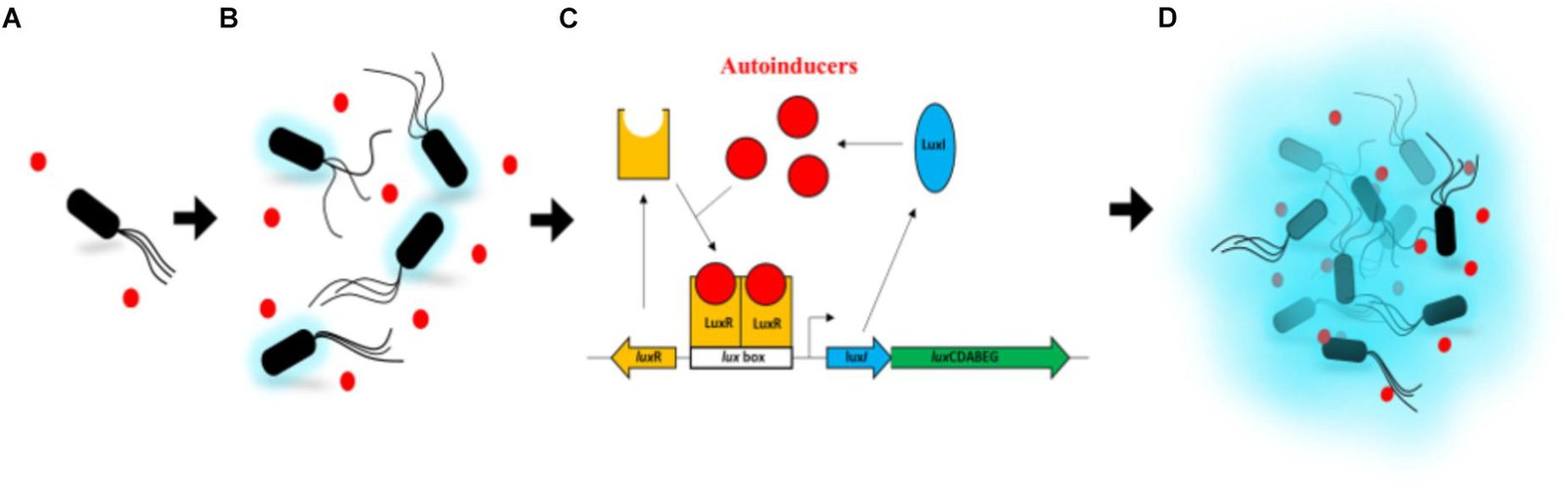
Some signal transduction pathways regulate the transcription of specific genes. This could be through the activation of transcription factors through phosphorylation or activation of second messengers. In quorum sensing, some autoinducers are hydrophobic in nature and will bind to cytoplasmic receptors (e.g., luxR). Once bound to its ligand, the luxR receptor can bind to specific sequences in the DNA and trigger transcription of both the luxR gene itself and the luxICDABEG operon (Figure 20.4.1). This results in higher expression of the luxR receptor (positive feedback) and expression of the proteins required to produce the luciferase protein. As expression of the lux operon increases, the levels of the protein luxI increases in the cytoplasm. luxI is an inhibitor of the autoinducer, which leads to the decrease of active luxR receptor promoting transcription. This ensures termination of the signal (negative feedback).
Increase in Cellular Metabolism
Signaling pathways can also influence cellular metabolism. This is primarily through enhancing or inhibiting the activities of metabolic proteins that are responsible for extracting energy from nutrients in the environment or modulate the ability to take up those same nutrients into the cell. A good example of this kind of signaling pathway that also involves transcriptional regulation, is the signaling pathway downstream of the lactose permease discussed in Section 19.1, Transcriptional Regulation of the lac Operon.
Cell Growth
Cell signaling pathways also play a major role in cell division. Cells do not normally divide unless they are stimulated by signals from other cells. The ligands that promote cell growth are called growth factors. Many growth factors bind to cell-surface receptors that are linked to tyrosine kinases. These cell-surface receptors are called receptor tyrosine kinases (RTKs). Activation of RTKs initiates a signaling pathway that then stimulates the expression of proteins that interact with other cellular components to initiate cell division.
Cell Death
When cells are damaged, unnecessary, or potentially harmful, they can undergo apoptosis, a form of programmed cell death that prevents the release of harmful contents. Apoptosis can be triggered internally by cell health checkpoints or externally by signals such as loss of contact with the extracellular matrix—a mechanism that helps prevent abnormal cell migration, like in cancer. In the immune system, apoptosis helps eliminate T-cells that mistakenly recognize the body’s own proteins, reducing the risk of autoimmune disease.
Apoptosis also plays a vital role in development. For instance, in vertebrate embryos, it removes the web-like tissue between developing fingers and toes, allowing for proper limb formation. This tightly regulated cell death process is essential for maintaining health, immune function, and normal development.
Termination of the Signal Cascade
The aberrant signaling often seen in tumor cells is proof that the termination of a signal at the appropriate time can be just as important as the initiation of a signal. One method of stopping a specific signal is to degrade the ligand or remove it so that it can no longer access its receptor. One reason that hydrophobic hormones like estrogen and testosterone trigger long-lasting events is because they bind carrier proteins. These proteins allow the insoluble molecules to be soluble in blood, but they also protect the hormones from degradation by circulating enzymes.
Inside the cell, many different enzymes reverse the cellular modifications that result from signaling cascades. For example, phosphatases are enzymes that remove the phosphate group attached to proteins by kinases in a process called dephosphorylation. Cyclic AMP (cAMP) is degraded into AMP by phosphodiesterase, and the release of calcium stores is reversed by the Ca2+ pumps that are located in the external and internal membranes of the cell.
Practice Questions
Glossary
apoptosis
programmed cell death
growth factor
ligand that binds to cell-surface receptors and stimulates cell growth
inhibitor
molecule that binds to a protein (usually an enzyme) and keeps it from functioning
phosphatase
enzyme that removes the phosphate group from a molecule that has been previously phosphorylated
phosphodiesterase
enzyme that degrades cAMP, producing AMP, to terminate signaling
Figure Descriptions
Figure 20.4.1 The image is a progression diagram with four labeled sections: A, B, C, and D, showing a bacterial communication process called quorum sensing. In section A, a single black rod-shaped bacterium with tail-like flagella is surrounded by a few small red circles representing molecules. Section B shows an increased number of similar bacteria and red molecules, with a light blue glow surrounding each bacterium, indicating activity. Section C is a schematic diagram detailing the molecular process involved in quorum sensing. It features red circles labeled “autoinducers” interacting with a yellow structure labeled “LuxR” and other components like “LuxI”, “lux box”, and an arrow labeled “luxCDABEG”. Section D depicts numerous bacteria and red circles emitting a stronger blue glow, suggesting a collective response. [Return to Figure 20.4.1]
Licenses and Attributions
“Response to the Signal” is adapted from “9.3 Response to the Signal” by Mary Ann Clark, Matthew Douglas, and Jung Choi for OpenStax Biology 2e used under CC BY 4.0. “Response to the Signal” is licensed under CC BY-NC-SA 4.0.
Media Attributions
- 1C_Activation_of_the_lux_operon_in_Aliivibrio_fischeri © Li, Z., and Nair, S. K. (2012). adapted by Lisa Tanet, Christian Tamburini, Chloé Baumas, Marc Garel, Gwénola Simon and Laurie Casalot is licensed under a CC BY-SA (Attribution ShareAlike) license
